

Big Data
Fraud Detection Course - 2020/2021
Nuno Moniz
nuno.moniz@fc.up.pt
Today
- Big Data (review)
- The data.table package
Handling Big Data in R
Big Data
What is Big Data?
Hadley Wickham (Chief Scientist at RStudio) In traditional analysis, the development of a statistical model takes more time than the calculation by the computer. When it comes to Big Data this proportion is turned upside down.
Wikipedia Collection of data sets so large and complex that it becomes difficult to process using on-hand database management tools or traditional data processing applications.
The 3 V’s Increasing volume (amount of data), velocity (speed of data in and out), and variety (range of data types and sources)
R and Big Data
- R keeps all objects in memory - potential problem for big data
- Still, current versions of R can address 8 TB of RAM on 64-bit machines
- Nevertheless, big data is becoming more and more a hot topic within the R community so new “solutions” are appearing!
Some rules of thumb
- Up to 1 million records - easy on standard R
- 1 million to 1 billion - possible but with additional effort
- More than 1 billion - possibly require map reduce algorithms that can be designed in R and processed with connectors to Hadoop and others
Big Data Approaches in R
- Reducing the dimensionality of data
- Get bigger hardware and/or parallelize your analysis
- Integrate R with higher performing programming languages
- Use alternative R interpreters
- Process data in batches
- Improve your knowledge of R and its inner workings / programming tricks
Get Bigger Hardware
- Buy more memory
- Buy better processing capabilities
- Multi-core, multi-processor, clusters
Some sources of extra information
Integrate R with higher performing programming languages
- R is very good at integrating easily with other languages
- You can easily do heavy computation parts in other language
- Still, this requires knowledge about these languages that may not be easily adaptable for data analysis tasks, in spite of their efficiency
Some sources of extra information
- The outstanding package Rcpp allows you to call C and C++ directly in the middle of R code D. Eddelbuettel (2013): Seamless R and C++ Integration with Rcpp. UserR! Series. Springer.
- Section 5 of the R manual “Writing R Extensions” talks about interfacing other languages
Use alternative R interpreters
- Some special-purpose R interpreters exist:
Process data in batches
- Store data on hard disk
- Load and process data in chuncks
- But, analysis has to be adapted to work by chunk, or methods have to be adapted to work with data types stored on hard disk
Some sources of extra information
Improve your knowledge of R and its inner workings/programming tricks
- Minimize copies of the data
- Hint: learn about the way R passes arguments to functions Outstanding source of information at http://adv-r.had.co.nz/memory.html of the book “Advanced R Programming” by Hadley Wickham
- Prefer integers over doubles when possible
- Only read the data you really need from files
- Use categorical variables (read factors in R) with care
- Use loops with care particularly if they are making copies of the data along their execution
Improve your knowledge of R and its inner workings/programming tricks
The following is strongly inspired by a Hadley Wickham talk
- The typical data analysis process
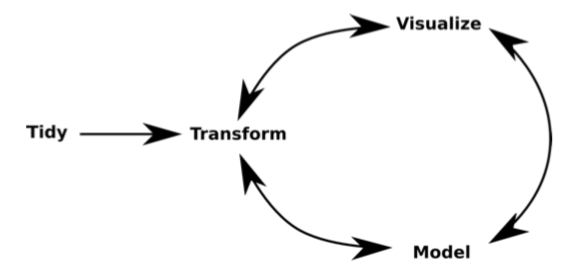
- On each of these steps there may be constraints with big data
Data Transformations
Split-Apply-Combine
- A frequent data transformation one needs to carry out
- Split the data set rows according to some criterion
- Calculate some value on each of the resulting subsets
- Aggregate the results into another aggregated data set
Split-Apply-Combine Example
library(plyr) # extra package you have to install data(algae, package="DMwR")
library(DMwR)
ddply(algae, .(season,speed), function(d) colMeans(d[, 5:7], na.rm=TRUE))
## season speed mnO2 Cl NO3
## 1 autumn high 11.145333 26.91107 5.789267
## 2 autumn low 10.112500 44.65738 3.071375
## 3 autumn medium 10.349412 47.73100 4.025353
## 4 spring high 9.690000 19.74625 2.013667
## 5 spring low 4.837500 69.22957 2.628500
## 6 spring medium 7.666667 76.23855 2.847792
## 7 summer high 10.629000 22.49626 2.571900
## 8 summer low 7.800000 58.74428 4.132571
## 9 summer medium 8.651176 47.23423 3.652059
## 10 winter high 9.760714 23.86478 2.738500
## 11 winter low 8.780000 43.13720 3.147600
## 12 winter medium 7.893750 66.95135 3.817609
- Nice and clean but slow on big data!
Remember dplyr?
dplyris a package by Hadley Wickham that re-invents several operations done withplyr(but) more efficiently
library(dplyr) # another extra package you have to install data(algae,package="DMwR")
grps <- group_by(algae, season, speed)
summarise(grps, avg.mnO2 = mean(mnO2, na.rm=TRUE),
avg.Cl = mean(Cl, na.rm = TRUE), avg.NO3 = mean(NO3, na.rm=TRUE))
## # A tibble: 12 x 5
## # Groups: season [4]
## season speed avg.mnO2 avg.Cl avg.NO3
## <fct> <fct> <dbl> <dbl> <dbl>
## 1 autumn high 11.1 26.9 5.79
## 2 autumn low 10.1 44.7 3.07
## 3 autumn medium 10.3 47.7 4.03
## 4 spring high 9.69 19.7 2.01
## 5 spring low 4.84 69.2 2.63
## 6 spring medium 7.67 76.2 2.85
## 7 summer high 10.6 22.5 2.57
## 8 summer low 7.8 58.7 4.13
## 9 summer medium 8.65 47.2 3.65
## 10 winter high 9.76 23.9 2.74
## 11 winter low 8.78 43.1 3.15
## 12 winter medium 7.89 67.0 3.82
Some comments on dplyr
- It is extremely fast and efficient
- It can handle not only data frames but also objects of class
data.tableand standard data bases - New developments may arise as it is a very new package
Data Visualization
- R has excellent facilities for visualizing data
- With big data plotting can become very slow
- Recent developments are trying to take care of this
- Hadley Wickham is developing a new package for this:
bigvis(here) - From the project page:
- The bigvis package provides tools for exploratory data analysis of large datasets (10-100 million obs).
- The aim is to have most operations take less than 5 seconds on commodity hardware, even for 100,000,000 data points.
Efforts on Modeling with Big Data
- Model construction with Big Data is particularly hard
- Most algorithms include sophisticated operations that frequently do not scale up very well
- The R community is making some efforts to alleviate this problem. A few examples:
bigrf- a package providing a Random Forests implementation with support for parallel execution and large memorybiglm,speedglm- packages for fitting linear and generalized linear models to large data
- A way to face the problem is through streaming algorithms
HadoopStreaming- Utilities for using R scripts in Hadoop streamingstream- interface to MOA open source framework for data stream mining
The data.table package
Package data.table
citation("data.table")
##
## To cite package 'data.table' in publications use:
##
## Matt Dowle and Arun Srinivasan (2020). data.table: Extension of
## `data.frame`. R package version 1.13.2.
## https://CRAN.R-project.org/package=data.table
##
## A BibTeX entry for LaTeX users is
##
## @Manual{,
## title = {data.table: Extension of `data.frame`},
## author = {Matt Dowle and Arun Srinivasan},
## year = {2020},
## note = {R package version 1.13.2},
## url = {https://CRAN.R-project.org/package=data.table},
## }
An extension of data.frame!
An extension of data.frame?!
library(data.table)
library(DMwR)
data(iris)
ds <- as.data.table(iris)
class(ds)
## [1] "data.table" "data.frame"
What is it?
Besides an R package ...
- (Overall) Fastest framework for data wrangling. And yes, it includes:
- plyr
- dplyr
- and pandas (Python)
If in doubt, check the benchmark by H2O.ai: https://h2oai.github.io/db-benchmark/
Goal 1 Reduce programming time (less function calls and name repetition)
Goal 2 Reduce compute time (faster aggregation and update by reference)
General idea: I-J-by
- DT[i, j, by]
i, j and by are the main components of data.table:
- i in R, is the WHERE in SQL;
- j is the SELECT, and;
- by is the GROUP BY.
Translating: Take DT, subset rows using i, then calculate j grouped by by
Basics: Data Selection
With data.frame...
ds[1:2, c("Sepal.Length","Species")]
## Sepal.Length Species
## 1: 5.1 setosa
## 2: 4.9 setosa
With data.table...
ds[1:2, list(Sepal.Length, Species)]
## Sepal.Length Species
## 1: 5.1 setosa
## 2: 4.9 setosa
Using .operation ...
ds[1:2, .(Sepal.Length, Species)]
## Sepal.Length Species
## 1: 5.1 setosa
## 2: 4.9 setosa
Data Filtering
You will save a lot of dollar signs.
With data.frame you need to specify the data structure in the conditions used
head(ds[ds$Sepal.Length>5, "Species"])
## Species
## 1: setosa
## 2: setosa
## 3: setosa
## 4: setosa
## 5: setosa
## 6: setosa
With data.table, the package makes that assumption.
head(ds[Sepal.Length>5, Species])
## [1] setosa setosa setosa setosa setosa setosa
## Levels: setosa versicolor virginica
Handling NA's
With data.frame you have to call the is.na() function
head(ds[!is.na(ds$Sepal.Length), "Species"])
## Species
## 1: setosa
## 2: setosa
## 3: setosa
## 4: setosa
## 5: setosa
## 6: setosa
With data.table, you can use na.omit directly
head(ds[, na.omit(Species)])
## [1] setosa setosa setosa setosa setosa setosa
## Levels: setosa versicolor virginica
New Columns?
In data.frame ...
ds["SepalRatio"] <- ds$Sepal.Length / ds$Sepal.Width
In data.table!
ds[, SepalRatio := Sepal.Length / Sepal.Width]
head(ds)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species SepalRatio
## 1: 5.1 3.5 1.4 0.2 setosa 1.457143
## 2: 4.9 3.0 1.4 0.2 setosa 1.633333
## 3: 4.7 3.2 1.3 0.2 setosa 1.468750
## 4: 4.6 3.1 1.5 0.2 setosa 1.483871
## 5: 5.0 3.6 1.4 0.2 setosa 1.388889
## 6: 5.4 3.9 1.7 0.4 setosa 1.384615
Remove a column!
In data.frame ...
ds$SepalRatio <- NULL
In data.table ... The syntax is not always easier.
ds[, SepalRatio := NULL]
head(ds)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1: 5.1 3.5 1.4 0.2 setosa
## 2: 4.9 3.0 1.4 0.2 setosa
## 3: 4.7 3.2 1.3 0.2 setosa
## 4: 4.6 3.1 1.5 0.2 setosa
## 5: 5.0 3.6 1.4 0.2 setosa
## 6: 5.4 3.9 1.7 0.4 setosa
Bonus: the .N
The .N operator references the last case in a data set.
It may (or may not) be useful in your endeavours with the PA.
ds[.N, ]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1: 5.9 3 5.1 1.8 virginica
Data Aggregation (Examples)
- Mean Sepal.Length by Species?
ds.aux <- ds[, mean(Sepal.Length), by=Species]
- Sorting (ascending and descending)
ds.aux <- ds.aux[order(V1)]
ds.aux
## Species V1
## 1: setosa 5.006
## 2: versicolor 5.936
## 3: virginica 6.588
ds.aux <- ds.aux[order(-V1)]
ds.aux
## Species V1
## 1: virginica 6.588
## 2: versicolor 5.936
## 3: setosa 5.006
The fread function (!)
If you have look at the data for the PA, you probably know that you will wait a bit to have it laoded into R.
data.table is going to be very useful in that aspect, thanks to fread()
set.seed() #input a random number of your choice
bulk <- fread("../../PA/Dataset/train.csv", sep=",", header=TRUE) # load the data set
ids <- sample(1:nrow(bulk), 0.1*nrow(bulk)) # select a sample (10%) of the entire data to speed up even further
ds <- bulk[ids,]
# Important note: if you're sampling, you should be considering the variance of results.
# This would mean that a single good result may not mean that what you are doing is best
Diving into data.table
- The previous slides provide a general overview of the syntax in data.table.
- Considering the size of the dataset you have for the PA, this could be a great tool.
- if you want to know more, there are great tutorials available:
Hands-on data.table
Hands-on data.table
Download the PA dataset
Load the data set using the fread function
Find out the percentage of fraud cases in the data set (isFraud feature)
Create a 10% sample of the training data set.
Create two data subsets: one for transactions carried out with a desktop and another with those performed with a mobile device (DeviceType). Calculate the percentage of fraud cases in each of these subsets (maybe the table function?)
Trim down the data subsets from the previous item by only selecting the following features: isFraud, TransactionDT, TransactionAmt, card1. Create a Random Forest model with the data subset concerning transactions made with desktop devices, and use this model to predict fraud in the transactions made by mobile devices.
Use the Metrics package and the auc function to calculate the Area Under the Curve.
